Finite Element Neural Network Interpolation: Hybridisation with the Proper Generalised Decomposition for Surrogate Modelling
DTE & AICOMAS 2025
LMS, École Polytechnique, Paris, France
February 21, 2025
Idiopathic Pulmonary Fibrosis
Objectives
- Improved understanding of the physiological mechanisms
- In silico prognostics
- Transfer tools to the clinic
- Patient-specific digital twins
Requirements
Mechanical model of the lung (Patte et al., 2022; Peyraut & Genet, 2024)
Uncertainty quantification (Laville et al., 2023; Peyraut & Genet, (In Revision))
Real-time estimation of the parameters
Lung poro-mechanics (Patte et al., 2022)
Weak formulation :
\[ \begin{cases} \int_{\Omega} \boldsymbol{\Sigma} : d_{\boldsymbol{u}}\boldsymbol{E}\left( \boldsymbol{u^*} \right) ~\mathrm{d}\Omega= W_{\text{ext}}\left(\boldsymbol{u},\boldsymbol{u^*}\right) ~\forall \boldsymbol{u^*} \in H^1_0\left(\Omega \right) \\[4mm] \int_{\Omega} \left(pf + \frac{\partial \Psi \left(\boldsymbol{u},\Phi_f\right)}{\partial \Phi_s}\right) \Phi_f^* ~\mathrm{d}\Omega= 0 ~\forall \Phi_f^* \end{cases} \label{eq:MechPb_lung} \]
Kinematics:
- \(\boldsymbol{F} = \boldsymbol{1} + \boldsymbol{\nabla}\boldsymbol{u}\) - deformation gradient
- \(\boldsymbol{C} = \boldsymbol{F}^T \cdot \boldsymbol{F}\) - right Cauchy-Green tensor
- \(\boldsymbol{E} = \frac{1}{2}\left(\boldsymbol{C} -\boldsymbol{1} \right)\) - Green-Lagrange tensor
- \(I_1 = \text{tr}\left(\boldsymbol{C} \right)\) - first invariant of \(\boldsymbol{C}\)
- \(I_2 = \frac{1}{2} \left(\text{tr}\left(\boldsymbol{C} \right)^2 - \text{tr}\left( \boldsymbol{C}^2 \right) \right)\)
Behaviour
- \(\boldsymbol{\Sigma} = \frac{\partial \Psi_s + \Psi_f}{\partial \boldsymbol{E}} = \frac{\partial \Psi_s }{\partial \boldsymbol{E}} + p_f J \boldsymbol{C}^{-1}\) - The second Piola-Kirchhoff stress tensor
- \(p_f = -\frac{\partial \Psi_s}{\partial \Phi_s}\) - fluid pressure
- \(\Psi_s\left(\boldsymbol{E}, \Phi_s \right) = W_{\text{skel}} + W_{\text{bulk}}\) \[\begin{cases} W_{\text{skel}} = \beta_1\left(I_1 - \text{tr}\left(I_d\right) -2 \text{ln}\left(\mathrm{det}\left(\boldsymbol{F} \right)\right)\right) + \beta_2 \left(I_2 -3 -4\text{ln}\left(\mathrm{det}\left(\boldsymbol{F} \right)\right)\right) + \alpha \left( e^{\delta\left(\mathrm{det}\left(\boldsymbol{F} \right)^2-1-2\text{ln}\left(\mathrm{det}\left(\boldsymbol{F} \right)\right) \right)} -1\right)\\ W_{\text{bulk}} = \kappa \left( \frac{\Phi_s}{1-\Phi_{f0}} -1 -\text{ln}\left( \frac{\Phi_s}{1-\Phi_{f0}}\right) \right) \end{cases}\]
- \(\Phi_s\) current volume fraction of solid pulled back on the reference configurations
Simpler mechanical problem - surrogate modelling
- Parametrised (patient-specific) mechanical problem
\[ \definecolor{BleuLMS}{RGB}{1, 66, 106} \definecolor{VioletLMS}{RGB}{77, 22, 84} \definecolor{TealLMS}{RGB}{0, 103, 127} % \definecolor{BleuLMS2}{RGB}{0, 103, 202} \definecolor{BleuLMS2}{RGB}{0, 169, 206} \definecolor{BleuLMPS}{RGB}{105, 144, 255} \definecolor{accentcolor}{RGB}{1, 66, 106} \definecolor{GreenLMS}{RGB}{0,103,127} \definecolor{LGreenLMS}{RGB}{67,176,42} \definecolor{RougeLMS}{RGB}{206,0,55} \]
\[ \boldsymbol{u} = \mathop{\mathrm{arg\,min}}_{H^1\left(\Omega \right)} \int_{\Omega} \Psi\left( \boldsymbol{E}\left(\boldsymbol{u}\left(\textcolor{BleuLMPS}{\boldsymbol{x}}, \textcolor{LGreenLMS}{\left\{\mu_i\right\}_{i \in \mathopen{~[\!\![~}1, \beta \mathclose{~]\!\!]}}}\right)\right) \right) ~\mathrm{d}\Omega- W_{\text{ext}}\left(\textcolor{BleuLMPS}{\boldsymbol{x}}, \textcolor{LGreenLMS}{\left\{\mu_i\right\}_{i \in \mathopen{~[\!\![~}1, \beta \mathclose{~]\!\!]}}}\right) \]
Kinematics:
- \(\boldsymbol{F} = \boldsymbol{1} + \boldsymbol{\nabla}\boldsymbol{u}\) - deformation gradient
- \(\boldsymbol{C} = \boldsymbol{F}^T \cdot \boldsymbol{F}\) - right Cauchy-Green tensor
- \(\boldsymbol{E} = \frac{1}{2}\left(\boldsymbol{C} -\boldsymbol{1} \right)\) - Green-Lagrange tensor
- \(I_1 = \text{tr}\left(\boldsymbol{C} \right)\) - first invariant of \(\boldsymbol{C}\)
Behaviour - Saint Venant-Kirchhoff
\(\Psi = \frac{\lambda}{2} \text{tr}\left(\boldsymbol{E}\right)^2 + \mu \boldsymbol{E}:\boldsymbol{E}\)
- Unstable under compression
- \(\nu = 0.49\) for quasi-incompressibility
- Soft tissues are often modelled as incompressible materials
Reduced-order model (ROM)
- Build a surrogate model
- Parametrised solution \(\left(\textcolor{BleuLMPS}{\boldsymbol{x}}, \textcolor{LGreenLMS}{\left\{\mu_i\right\}_{i \in \mathopen{~[\!\![~}1, \beta \mathclose{~]\!\!]}}}\right)\)
- No computations online
- On the fly evaluation of the model
- Real-time parameters estimation
Outline
|
|
II - Methods
\[ \definecolor{BleuLMS}{RGB}{1, 66, 106} \definecolor{VioletLMS}{RGB}{77, 22, 84} \definecolor{TealLMS}{RGB}{0, 103, 127} % \definecolor{BleuLMS2}{RGB}{0, 103, 202} \definecolor{BleuLMS2}{RGB}{0, 169, 206} \definecolor{BleuLMPS}{RGB}{105, 144, 255} \definecolor{accentcolor}{RGB}{1, 66, 106} \definecolor{GreenLMS}{RGB}{0,103,127} \definecolor{LGreenLMS}{RGB}{67,176,42} \definecolor{RougeLMS}{RGB}{206,0,55} \]
|
|
|
|
Interpolation of the solution
- FEM finite admissible space
\[ \mathcal{U}_h = \left\{\boldsymbol{u}_h \; | \; \boldsymbol{u}_h \in \text{Span}\left( \left\{ N_i^{\Omega}\left(\boldsymbol{x} \right)\right\}_{i \in \mathopen{~[\!\![~}1,N\mathclose{~]\!\!]}} \right)^d \text{, } \boldsymbol{u}_h = \boldsymbol{u}_d \text{ on }\partial \Omega_d \right\} \]
Interpretabilty Kronecker property Easy to prescribe Dirichlet boundary conditions
Mesh adaptation not directly embedded in the method
- Physics Informed Neural Networks (PINNs)
\[ \boldsymbol{u} \left(x_{0,0,0} \right) = \sum\limits_{i = 0}^C \sum\limits_{j = 0}^{N_i} \sigma \left( \sum\limits_{k = 0}^{M_{i,j}} b_{i,j}+\omega_{i,j,k}~ x_{i,j,k} \right) \]
All interpolation parameters are trainable Benefits from ML developments
Not easily interpretable Difficult to prescribe Dirichlet boundary conditions
- HiDeNN framework (Liu et al., 2023; Park et al., 2023; Zhang et al., 2021)
- Best of both worlds
- Reproducing FEM interpolation with constrained sparse neural networks
- \(N_i^{\Omega}\) are SNN with constrained weights and biases
- Fully interpretable parameters
- Continuous interpolated field that can be automatically differentiated
- Runs on GPUs
Solving the mechanical problem
Solving the mechanical problems amounts to finding the continuous displacement field minimising the potential energy \[E_p\left(\boldsymbol{u}\right) = \frac{1}{2} \int_{\Omega}\boldsymbol{E} : \mathbb{C} : \boldsymbol{E} ~\mathrm{d}\Omega- \int_{\partial \Omega_N}\boldsymbol{F}\cdot \boldsymbol{u} ~\mathrm{d}A- \int_{\Omega}\boldsymbol{f}\cdot\boldsymbol{u}~\mathrm{d}\Omega. \]
In practice
- Compute the loss \(\mathcal{L} := E_p\)
- Find the parameters minimising the loss
- Rely on state-of-the-art optimizers (ADAM, LBFGS, etc.)
Degrees of freedom (Dofs)
- Nodal values (standard FEM Dofs)
- Nodal coordinates (Mesh adaptation)
- hr-adaptivity
h-adaptivity: red-green refinement
technical details:
| Dofs (initially) | \(569~438\) |
|---|---|
| Training time (s) | \(7\) |
| GPU (V100) | 1 |
| CPUs | 40 |
Reduced-order modelling (ROM)
Low-rank approximation of the solution to avoid the curse of dimensionality with \(\beta\) parameters
Full-order discretised model
- \(N\) spatial shape functions
- Span a finite spatial space of dimension \(N\)
- Requires computing \(N\times \beta\) associated parametric functions
Reduced-order model
- \(m \ll N\) spatial modes
- Similar to global shape functions
- Span a smaller space
- Galerkin projection
Finding the reduced-order basis
- Linear Normal Modes (eigenmodes of the structure) (Hansteen & Bell, 1979)
- Do not account for the loading and behaviour specifics
- Proper Orthogonal Decomposition (Chatterjee, 2000; Radermacher & Reese, 2013)
- Require wise selection of the snapshots and costly computations
- Reduced basis method (Maday & Rønquist, 2002) with EIM (Barrault et al., 2004)
- Rely on prior expensive computations
Proper Generalised Decomposition (PGD)
PGD: (Chinesta et al., 2011; Ladeveze, 1985)
Tensor decomposition
- Separation of variables
- Low-rank \(m\)
- \(\textcolor{BleuLMPS}{\overline{\boldsymbol{u}}_i(\boldsymbol{x})}\) Space modes
- \(\textcolor{LGreenLMS}{\lambda_i^j(\mu^j)}\) Parameter modes
\[\textcolor{VioletLMS}{\boldsymbol{u}}\left(\textcolor{BleuLMPS}{\boldsymbol{x}}, \textcolor{LGreenLMS}{\mu}\right) = \sum\limits_{i=0}^{m}\textcolor{BleuLMPS}{\overline{u}_i\left(x\right)}\textcolor{LGreenLMS}{\lambda_i\left(\mu\right)}\]
Discretised problem
- From \(N\times N_{\mu}\) unknowns to \(m\times\left(N + N_{\mu}\right)\)
- e.g. \(10000 \times 1000 = 1e7 \gg 5\left( 10 000+ 1000 \right) = 5.5e4\)
Proper Generalised Decomposition (PGD)
PGD: (Chinesta et al., 2011; Ladeveze, 1985)
Tensor decomposition - extra-coordinates
- Separation of variables
- Low-rank \(m\)
- \(\textcolor{BleuLMPS}{\overline{\boldsymbol{u}}_i(\boldsymbol{x})}\) Space modes
- \(\textcolor{LGreenLMS}{\lambda_i^j(\mu^j)}\) Parameter modes
\[ \textcolor{VioletLMS}{\boldsymbol{u}}\left(\textcolor{BleuLMPS}{\boldsymbol{x}}, \textcolor{LGreenLMS}{\left\{\mu_i\right\}_{i \in \mathopen{~[\!\![~}1, \beta \mathclose{~]\!\!]}}}\right) = \sum\limits_{i=1}^m \textcolor{BleuLMPS}{\overline{\boldsymbol{u}}_i(\boldsymbol{x})} ~\textcolor{LGreenLMS}{\prod_{j=1}^{\beta}\lambda_i^j(\mu^j)} \]
Discretised problem
- From \(N\times\prod\limits_{j=1}^{~\beta} N_{\mu}^j\) unknowns to \(m\times\left(N + \sum\limits_{j=1}^{\beta} N_{\mu}^j\right)\)
- e.g. \(10000 \times 1000^2 = 1e10 \gg 5\left( 10 000+ 2\times 1000 \right) = 6e4\)
- Finding the tensor decomposition by minimising the energy \[ \left(\left\{\overline{\boldsymbol{u}}_i \right\}_{i\in \mathopen{~[\!\![~}1,m\mathclose{~]\!\!]}},\left\{\lambda_i^j \right\}_{ \begin{cases} i\in \mathopen{~[\!\![~}1,m\mathclose{~]\!\!]}\\ j\in \mathopen{~[\!\![~}1,\beta \mathclose{~]\!\!]} \end{cases} } \right) = \mathop{\mathrm{arg\,min}}_{ \begin{cases} \left(\overline{\boldsymbol{u}}_1, \left\{\overline{\boldsymbol{u}}_i \right\} \right) & \in \mathcal{U}\times \mathcal{U}_0 \\ \left\{\left\{\lambda_i^j \right\}\right\} & \in \left( \bigtimes_{j=1}^{~\beta} \mathcal{L}_2\left(\mathcal{B}_j\right) \right)^{m-1} \end{cases}} ~\underbrace{\int_{\mathcal{B}}\left[E_p\left(\boldsymbol{u}\left(\boldsymbol{x},\left\{\mu_i\right\}_{i \in \mathopen{~[\!\![~}1, \beta \mathclose{~]\!\!]}}\right), \mathbb{C}, \boldsymbol{F}, \boldsymbol{f} \right) \right]\mathrm{d}\beta}_{\mathcal{L}} \label{eq:min_problem} \]
Proper Generalised Decomposition (PGD)
- Building the low-rank tensor decomposition greedily (Ladeveze, 1985; Nouy, 2010)
\[ \boldsymbol{u}\left(\boldsymbol{x}, \left\{\mu_i\right\}_{i \in \mathopen{~[\!\![~}1, \beta \mathclose{~]\!\!]}}\right) = \overline{\boldsymbol{u}}(\boldsymbol{x}) ~\prod_{j=1}^{\beta}\lambda^j(\mu^j) \]
\[ \boldsymbol{u}\left(\boldsymbol{x}, \left\{\mu_i\right\}_{i \in \mathopen{~[\!\![~}1, \beta \mathclose{~]\!\!]}}\right) = \textcolor{RougeLMS}{\sum\limits_{i=1}^{2}} \overline{\boldsymbol{u}}_{\textcolor{RougeLMS}{i}}(\boldsymbol{x}) ~\prod_{j=1}^{\beta}\lambda_{\textcolor{RougeLMS}{i}}^j(\mu^j) \]
\[ \boldsymbol{u}\left(\boldsymbol{x}, \left\{\mu_i\right\}_{i \in \mathopen{~[\!\![~}1, \beta \mathclose{~]\!\!]}}\right) = \sum\limits_{i=1}^{\textcolor{RougeLMS}{m}} \overline{\boldsymbol{u}}_i(\boldsymbol{x}) ~\prod_{j=1}^{\beta}\lambda_i^j(\mu^j) \]
Greedy algorithm
- Start with a single mode
- Minimise the loss until stagnation
- Add a new mode
- If loss decreases, continue training
- Else stop and return the converged model
The PGD is
- An a priori ROM techniques
- Building the model from scratch on the fly
- No full-order computations
- Reduced-order basis tailored to the specifics of the problem (Daby-Seesaram et al., 2023)
Note
- Training of the full tensor decomposition, not only the \(m\)-th mode
- Actual minimisation implementation of the PGD
- No need to solve a new problem for any new parameter
- The online stage is a simple evaluation of the tensor decomposition
Neural Network PGD
Graphical implementation of Neural Network PGD
\[ \boldsymbol{u}\left(\textcolor{BleuLMPS}{\boldsymbol{x}}, \textcolor{LGreenLMS}{\left\{\mu_i\right\}_{i \in \mathopen{~[\!\![~}1, \beta \mathclose{~]\!\!]}}}\right) = \sum\limits_{i=1}^m \textcolor{BleuLMPS}{\overline{\boldsymbol{u}}_i(\boldsymbol{x})} ~\textcolor{LGreenLMS}{\prod_{j=1}^{\beta}\lambda_i^j(\mu^j)} \]
Interpretable NN-PGD
- No black box
- Fully interpretable implementation
- Great transfer learning capabilities
- Straightforward implementation
- Benefiting from current ML developments
- Straightforward definition of the physical loss using auto-differentiation
Stiffness and external forces parametrisation
Illustration of the surrogate model in use (preprint: Daby-Seesaram et al., 2024)
Parameters
- Parametrised stiffness \(E\)
- Parametrised external force \(\boldsymbol{f} = \begin{bmatrix} ~ \rho g \sin\left( \theta \right) \\ -\rho g \cos\left( \theta \right) \end{bmatrix}\)
NN-PGD
- Immediate evaluation of the surrogate model
- Straightforward differentiation capabilities regarding the input parameters
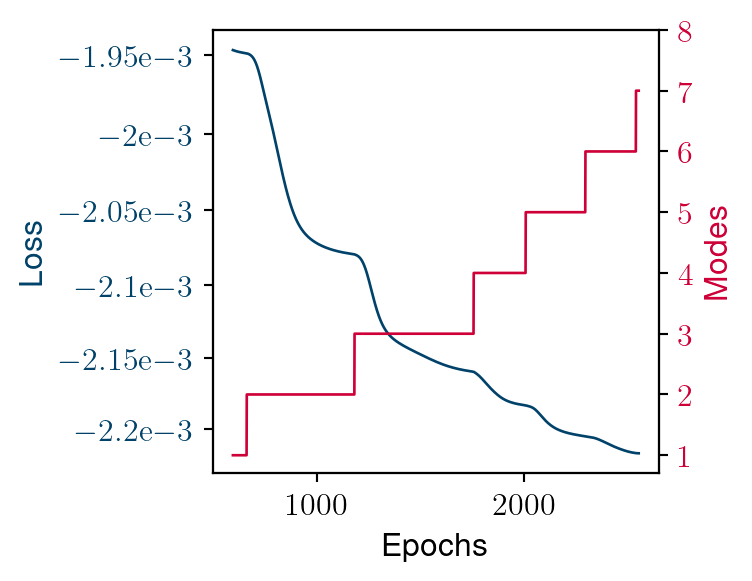
Convergence of the greedy algorithm
Loss convergence
Loss decay
Construction of the ROB
- New modes are added when the loss’s decay cancels out
- Adding a new mode gives extra latitude to improve predictivity
Accuracy with regards to FEM solutions
| Solution | Error |
|---|---|
\(E = 3.8 \times 10^{-3}\mathrm{ MPa}, \theta = 241^\circ\) |
\(E = 3.8 \times 10^{-3}\mathrm{ MPa}, \theta = 241^\circ\) |
\(E = 3.1 \times 10^{-3}\mathrm{ MPa}, \theta = 0^\circ\) |
\(E = 3.1 \times 10^{-3}\mathrm{ MPa}, \theta = 0^\circ\) |
| \(E\) (kPa) | \(\theta\) (rad) | Relative error |
|---|---|---|
| 3.80 | 1.57 | \(1.12 \times 10^{-3}\) |
| 3.80 | 4.21 | \(8.72 \times 10^{-4}\) |
| 3.14 | 0 | \(1.50 \times 10^{-3}\) |
| 4.09 | 3.70 | \(8.61 \times 10^{-3}\) |
| 4.09 | 3.13 | \(9.32 \times 10^{-3}\) |
| 4.62 | 0.82 | \(2.72 \times 10^{-3}\) |
| 5.01 | 2.26 | \(5.35 \times 10^{-3}\) |
| 6.75 | 5.45 | \(1.23 \times 10^{-3}\) |
Multi-level training
Strategy
- The interpretability of the NN allows
- Great transfer learning capabilities
- Start with coarse (cheap) training
- Followed by fine tuning, refining all modes
- Great transfer learning capabilities
- Extends the idea put forward by (Giacoma et al., 2015)
Note
- The structure of the Tensor decomposition is kept throughout the multi-level training
- Last mode of each level is irrelevant by construction
- It is removed before passing onto the next refinement level
III - Patient-specific parametrisation
|
|
Shape model - registration (Gangl et al., 2021)
Computing the mappings
- The objective is to find the domain \(\omega\) occupied by a given segmented shape in a 3D image
- Which is solution of
\[\omega = \arg \min\underbrace{ \int_{\omega} I\left( x \right) \mathrm{d}\omega}_{\Psi\left(\omega\right)}\]
With requirement that \(I\left( x \right) \begin{cases} < 0 \forall x \in \mathcal{S} \\ > 0 \forall x \not \in \mathcal{S} \end{cases}\), with \(\mathcal{S}\) the image footprint of the shape to be registered
The domain \(\omega\) can be described by mapping \[\Phi\left(X\right) = I_d + u\left(X\right)\] from a reference shape \(\Omega\), leading to \[\int_{\omega} I\left( x \right) \mathrm{d}\omega = \int_{\Omega} I \circ \Phi \left( X \right) J \mathrm{d}\Omega\]
Shape derivatives
- \(D\Psi\left(\omega\right)\left(\boldsymbol{u}^*\right) = \int_{\Omega} \nabla I \cdot u^* J \mathrm{d}\Omega + \underbrace{\int_{\Omega}I \circ \Phi\left(X\right) (F^T)^{-1} : \nabla u^* J \mathrm{d}\Omega}_{\int_{\omega}I\left(x\right) \mathrm{div}\left( u^* \circ \Phi \right) \mathrm{d}\omega}\)
Sobolev gradient (Neuberger, 1985; Neuberger, 1997)
\[D\Psi\left(u\right)\left(v^*\right) = \left(\nabla^H \Psi \left(u\right), v^* \right)_H\]
- Classically:
- \(\left(\boldsymbol{u}, \boldsymbol{v}^* \right)_H := \int_{\omega} \boldsymbol{\nabla_s}\boldsymbol{u}:\boldsymbol{\nabla_s}\boldsymbol{\boldsymbol{v}^*}\mathrm{d}\omega + \alpha \int_{\omega} \boldsymbol{u} \cdot \boldsymbol{v^*}\mathrm{d}\omega\)
Shape model - parametrisation
Encoding the shape of the lung in low-dimensional spaces
- in order to feed the surrogate model we need a parametrisation of the lung
- ROM on the shape mappings librairy
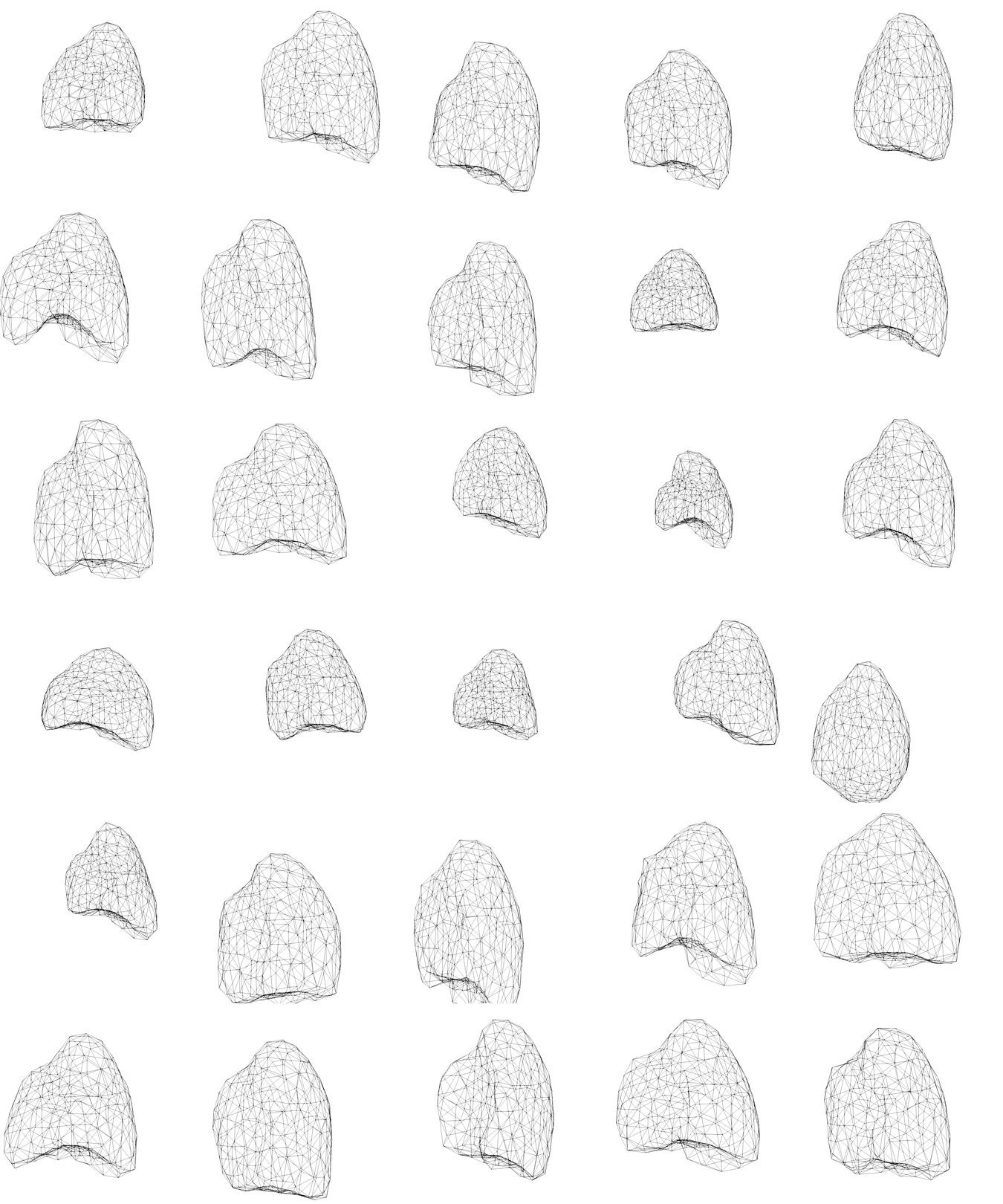
Shape model - parametrisation
Encoding the shape of the lung in low-dimensional spaces
- in order to feed the surrogate model we need a parametrisation of the lung
- ROM on the shape mappings librairy
First investigations - SVD
- ~40 patients
- SVD shows a relatively quick decay of the singular values
- Could benefit from non-linear reduction
- k-PCA in perspective
- possibility to use autoencoder
Conclusion
Conclusion
- Robust and straightforward general implementation of a NN-PGD
- Interpretable
- Benefits from all recent developments in machine learning
- Surrogate modelling of parametrised PDE solution
- Promising results on simple toy case
Perspectives for patient-specific applications
- Implementation of the poro-mechanics lung model (Patte et al., 2022)
- Further work on parametrising the geometries
- Inputting geometry parameters in the NN-PGD
- Error quantification on the estimated parameters
Technical perspectives
- 3D implementation
- Seamless coupling of both tools
- Do not hesitate to check the Github page
References
Appendix - Orthonormalisation of the ROB
- To ensure orthonormality of the RON, a Gramm-Schmidt like algorithm can be performed
\[ \boldsymbol{u}_{m+1} = \sum_{i=1}^{m} \underbrace{(\lambda_i + \lambda_{m+1} ~ \overline{\boldsymbol{u}}_i^T \overline{\boldsymbol{u}})}_{\mathring{\lambda}_i} \overline{\boldsymbol{u}}_i + \lambda_{m+1} \underbrace{\left(\overline{\boldsymbol{u}}_{m+1} - \sum_{i=1}^{m} ( \overline{\boldsymbol{u}}_i^T \overline{\boldsymbol{u}}) \overline{\boldsymbol{u}}_i\right)}_{\mathring{\overline{\boldsymbol{u}}}_{m+1}} \]
\[\overline{\boldsymbol{u}}_{m+1} \longleftarrow \frac{\mathring{\overline{\boldsymbol{u}}}_{m+1}}{\Vert\mathring{\overline{\boldsymbol{u}}}_{m+1} \Vert_{\mathbb{K}}} \]
\[ \lambda_{m+1} \longleftarrow \lambda_{m+1} \Vert\mathring{\overline{\boldsymbol{u}}}_{m+1} \Vert_{\mathbb{K}} \] \[ \{\lambda_i\}_{i\in\mathopen{~[\!\![~}1,m\mathclose{~]\!\!]}} \gets \{\mathring{\lambda}_i\}_{i\in\mathopen{~[\!\![~}1,m\mathclose{~]\!\!]}} \]
- Much more tricky with more thant 2 parameters in the tensor decomposition
Appendix - PGD of a known field & eigen value problem
- Let \(\boldsymbol{u}\left(\boldsymbol{x},t\right)\) defined on \(t \in I = \left[0,T\right]\) and \(\boldsymbol{x} \in \Omega \subset \mathbb{R}^d\). Let’s look at the single mode decomposition: \[\left(\boldsymbol{\overline{u}}_1, \lambda_1\right) = \mathop{\mathrm{arg\,min}}_{\mathcal{U},\mathcal{I}} \left\Vert \boldsymbol{u} - \boldsymbol{\overline{u}}_1 \lambda_1 \right\Vert^2\]
\[\Leftrightarrow\int_{I\times\Omega}\left(\boldsymbol{u} - \boldsymbol{\overline{u}}_1 \lambda_1 \right)\left( \boldsymbol{\overline{u}}^* \lambda^* \right) ~\mathrm{d}\Omega\mathrm{d}t = 0 \forall \lambda^* \in \mathcal{I}, \forall \boldsymbol{\overline{u}}^* \in \mathcal{U}\]
\[\Leftrightarrow\begin{cases} \lambda_1\left(t \right) = \frac{\int_{\Omega}\boldsymbol{u}\left(\boldsymbol{x},t \right)\boldsymbol{\overline{u}}_1 ~\mathrm{d}\Omega}{\int_{\Omega} \boldsymbol{\overline{u}}_1^2 ~\mathrm{d}\Omega} = f\left( \lambda_1 \right) \\ \boldsymbol{\overline{u}}_1\left(\boldsymbol{x} \right) =\frac{\int_{I}\boldsymbol{u}\left(\boldsymbol{x},t \right) \lambda_1 \mathrm{d}t}{\int_{I} \lambda_1^2 \mathrm{d}t} = g\left( \boldsymbol{\overline{u}}_1 \right) \end{cases}\]
\[ \Leftrightarrow \lambda_1 = f \circ g \left( \lambda_1 \right)\]
\[ \Leftrightarrow \underbrace{\int_{\Omega} \boldsymbol{u}\int_{I}\boldsymbol{u}\lambda_1 \mathrm{d}t ~\mathrm{d}\Omega}_{T\left(\lambda_1 \right)} = \underbrace{\frac{\int_{\Omega \left(\int_I \boldsymbol{u}\lambda_1\mathrm{d}t \right)^2 ~\mathrm{d}\Omega}}{\int_I \lambda_1^2\mathrm{d}t}}_ {\omega^2}\lambda_1\]
- \(\left\Vert \boldsymbol{u} - \boldsymbol{\overline{u}}_1\lambda_1\right\Vert = \left\Vert \boldsymbol{u}\right\Vert^2 - R\left( \lambda_1 \right)\)
with
\(R\left( \lambda_1 \right) = \omega^2\)
Eigen value problem
- Equivalent to searching for the stationarity of a rayleigh’s quotient
- It can be shown that minimising the truncation error amounts to fining the largest eigenvalues
In practice
- Modes are computed greedily
- Less optimal
- The temporal modes can be updated
- improved accuracy
Appendix - Einstein notation
Writing the linear elasticity with the einstein notation:
- \[ W_{\text{int}} = \int_{\mathcal{B}}\frac{1}{2} \int_{\Omega}\mathcal{\boldsymbol{\varepsilon}}: \mathbb{C} : \mathcal{\boldsymbol{\varepsilon}}~\mathrm{d}\Omega\mathrm{d}\beta = \frac{1}{2} K_{ij} \mathcal{\boldsymbol{\varepsilon}}_{j}\left(\boldsymbol{\overline{u}}_{em}\right) \mathcal{\boldsymbol{\varepsilon}}_{i}\left(\boldsymbol{\overline{u}}_{el}\right) \mathrm{det}\left(J\right)_{em} \lambda^{\left(1\right)}_{mp} \lambda^{\left(2\right)}_{lp} \lambda^{\left(1\right)}_{mt} \lambda^{\left(2\right)}_{lt} E_p\]
Appendix - Summary of the framework
Appendix - Curse of dimensionality in computing the loss
Computing the non-linear loss using einsum relies on the tensor decomposition and broadcasting to remain efficient
Although the displacement is represented through a tensor decomposition \(\boldsymbol{u}\left(\boldsymbol{x}, \left\{\mu_i\right\}_{i \in \mathopen{~[\!\![~}1, \beta \mathclose{~]\!\!]}}\right) = \boldsymbol{\overline{u}}_i \prod_{j=1}^{\beta}\lambda_i^j(\mu^j)\), some non-linear mapping of that function cannot be easily computed on the independent modes but requires building the full-order solution tensor which leads to prohibitive training costs.
More physically sound behaviour laws
- Incompressible Neo-Hookean elastic behaviour: Improve the elastic behaviour \[ \begin{cases} \Psi = C_1(\text{tr}\left(\boldsymbol{C}-\boldsymbol{1}\right)) + f\left(\text{ln}\left(\text{det}\left( \boldsymbol{F}\right) \right) \right)\\[2mm] \left(\text{det}\boldsymbol{F} - 1\right)^2 = 0 \end{cases} \]
Sparse sampling of the hypercube
- One idea might be to look at sparse sampling of the parameters sets over which the loss function is computed, using a (greedy) LHS in the parametric space for instance instead of computing the integral of the potential energy over the full parametric hypercube
- Similar idea than in (lu_adaptive_2018?) but applied on PDEs and not data compression
- The underlying idea being that the solution interpolation being constrained to be low-rank by the TD, such sparse training might suffice
Heterogeneous stiffness
- Bi-stiffness structure with unknown
- Stiffness jump \(\Delta E\)
- Jump position \(\alpha\)
- Towards the parametrisation of the disease’s progression
- Can be adapted to different stages of the diseases
- Different physiological initial stiffness
Heterogeneous stiffness
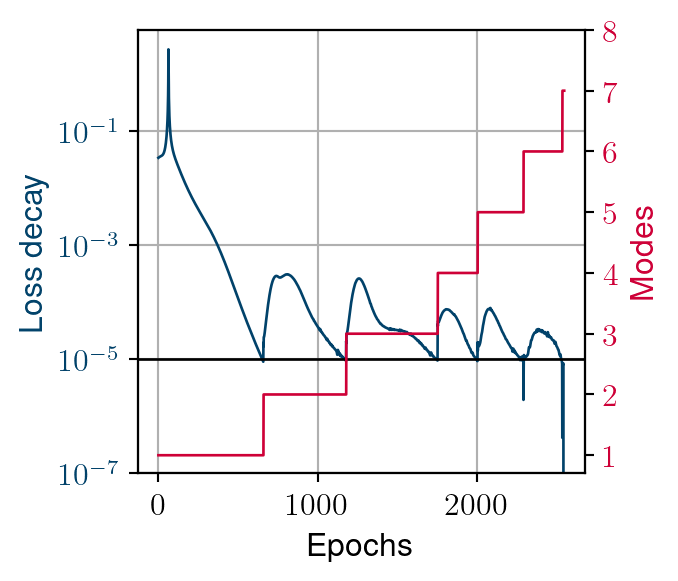
Convergence process
- Larger Kolmogorov n-with’s example
- Similar behaviour in constructing the reduced-order basis
- Adding a mode leads to a recovery in loss decay
- This recovery decreases until global stagnation
Hybridising ROM with NN
Shape model - registration (Gangl et al., 2021)
Minimisation problem
- The objective is to find the domain \(\omega\) occupied by a given segmented shape in a 3D image
- Which is solution of
\[\omega = \arg \min\underbrace{ \int_{\omega} I\left( x \right) \mathrm{d}\omega}_{\Psi\left(\omega\right)}\]
With requirement that \(I\left( x \right) \begin{cases} < 0 \forall x \in \mathcal{S} \\ > 0 \forall x \not \in \mathcal{S} \end{cases}\), with \(\mathcal{S}\) the image footprint of the shape to be registered
The domain \(\omega\) can be described by mapping \[\Phi\left(X\right) = I_d + u\left(X\right)\] from a reference shape \(\Omega\), leading to \[\int_{\omega} I\left( x \right) \mathrm{d}\omega = \int_{\Omega} I \circ \Phi \left( X \right) J \mathrm{d}\Omega\]
Shape derivatives
\(D\Psi\left(\omega\right)\left(\boldsymbol{u}^*\right) = \int_{\Omega} \nabla I \cdot u^* J \mathrm{d}\Omega + \underbrace{\int_{\Omega}I \circ \Phi\left(X\right) (F^T)^{-1} : \nabla u^* J \mathrm{d}\Omega}_{\int_{\omega}I\left(x\right) \mathrm{div}\left( u^* \circ \Phi \right) \mathrm{d}\omega}\)
Note that \[\int_{\omega}I\left(x\right) \mathrm{div}\left( u^* \circ \Phi \right) \mathrm{d}\omega = \int_{\partial \omega} I u^* \cdot n \partial \omega\]
Flat parts of the image result in non-zero derivative of the loss - The border is “pushed in the right direction”
Sobolev gradient (Neuberger, 1985; Neuberger, 1997)
- The regularisation is added through the choice of inner product \(H\) to reconstruct the gradient
\[D\Psi\left(u\right)\left(v^*\right) = \left(\nabla^H \Psi \left(u\right), v^* \right)_H\]
- Classically:
- \(\left(\boldsymbol{u}, \boldsymbol{v}^* \right)_H := \int_{\omega} \boldsymbol{\nabla_s}\boldsymbol{u}:\boldsymbol{\nabla_s}\boldsymbol{\boldsymbol{v}^*}\mathrm{d}\omega + \alpha \int_{\omega} \boldsymbol{u} \cdot \boldsymbol{v^*}\mathrm{d}\omega\)


